Workshop
Workshop Objectives
ABOUT WORKSHOP
Key components of the workshop include
An introduction to molecular docking tools such as AutoDock, AMDOCK, and CBDOCK. Practical exercises in docking small molecules into protein targets. Protein-protein docking using ClusPro and Rosetta Server. Fundamentals of molecular dynamics (MD) simulations with NAMD and GROMACS, including the analysis of MD simulation results. The role of bioinformatics in deciphering disease mechanisms. Systems biology approaches to drug discovery.


Learning Outcomes
Upon completing this workshop, participants will be able to
1. Critically evaluate the results of molecular docking and simulations, thereby optimizing the design and development of therapeutic agents and enhancing the probability of successful drug development.
2. Attain a comprehensive understanding of the fundamentals of drug design using computational approaches.
3. Achieve proficiency in structure- and ligand-based drug design techniques to develop potent drug candidates
4. Develop expertise in refinement of molecular models, performing simulations, and interpreting results to assess the behavior and properties of drug candidates.
5.Generative AI/ Machine Learning Based Drug Design.
CONVENOR

Prof. Nasimul Hoda
Jamia Millia Islamia,New Delhi
(National PI)

Prof. J. L. Vennerstrom
The University of Nebraska Medical Center USA
(International Co- PI)

Prof. Apurba Dutta
The University of Kansas, USA
(International PI)

Prof. B. Jayaram
Indian Institute of Technology, Delhi
(National Co-P1)
All participants are expected to come away with the basics about Docking and MD Simulation
Day-to-Day Programme



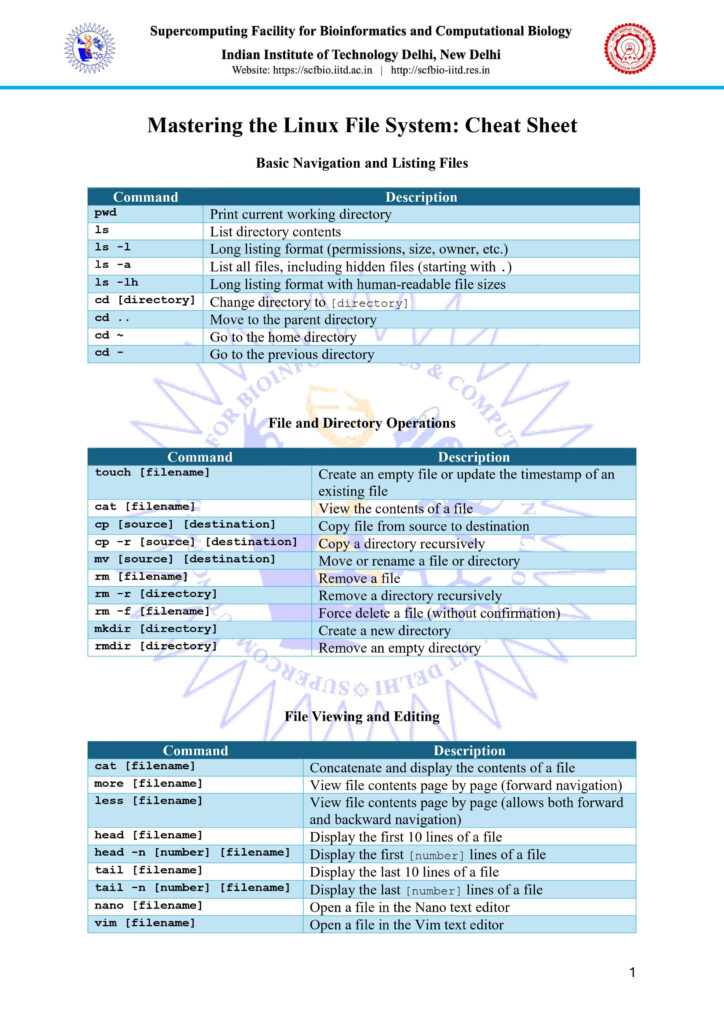
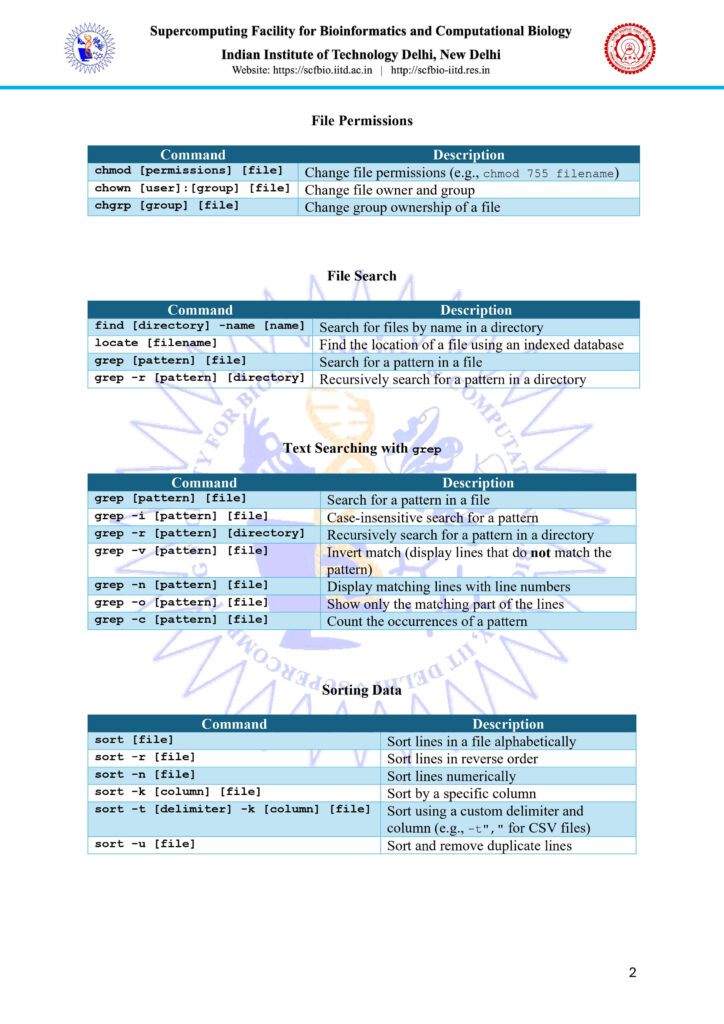
Important Linux Commands

Eligibility to Participate
Students of Postgraduate Onwards, Researchers, Working Professionals in Drug Design and Medicinal Chemistry or Allied Areas, and Young Faculty Members.
Venue: FTK-CIT Conference Hall, Jamia Millia Islamia, New Delhi